The mechanisms of normal division in a synthetic cell have proven hard to replicate in vitro, but one team of scientists has made a breakthrough that could be fundamental to the field of cell engineering.
Five years ago, scientists created a single-celled synthetic organism that, with only 473 genes, was the simplest living cell ever known.
However, this bacteria-like organism behaved strangely when growing and dividing, producing cells with wildly different shapes and sizes.
Now, a collaborative team from the J. Craig Venter Institute (JCVI), the National Institute of Standards and Technology (NIST) and the Massachusetts Institute of Technology (MIT) Center for Bits and Atoms, have identified seven genes that can be added to tame the cells’ unruly nature, causing them to neatly divide into uniform orbs.
According to the researchers, identifying these genes is an important step toward engineering synthetic cells that have useful medical applications, like acting as small factories that produce drugs, foods and fuels, or detecting diseases and producing drugs to treat them while living inside the body.
“Designing whole genomes to achieve desired phenotypes stands as a grand challenge in synthetic biology.
“But our capacity to synthesise and modify genomes has rapidly outpaced our ability to predict phenotype from genotype for large-scale genome design.
“Our work uses reverse genetics to understand the function of genes of unknown function involved in the basic cell processes of controlling cell size and shape, and cell division.
“Every gene we are able to pair with its function gets us closer to realising the goal of designing genomes for engineering cells.”
The full study published to document the team’s findings can be found in the journal Cell.
How did the researchers discover the mechanisms for synthetic cell division?
Trial and error
Scientists at JCVI constructed the first cell with a synthetic genome in 2010.
They didn’t build that cell completely from scratch, but started with cells from a very simple type of bacteria called a mycoplasma.
They destroyed the DNA in those cells and replaced it with DNA that was designed on a computer and synthesised in a lab.
This was the first organism in the history of life on Earth to have an entirely synthetic genome – they called it JCVI-syn1.0.
Since then, scientists have been working to strip that organism down to its minimum genetic components.
But according to the researchers, the super-simple cell JCVI created five years ago, dubbed JCVI-syn3.0, was perhaps too minimalist.
The collaborative team added 19 genes back to this cell, including the seven needed for normal cell division, to create the new variant – JCVI-syn3A.
This variant has fewer than 500 genes. To put that number into perspective, the E. coli bacteria that live in the human gut have about 4,000 genes, and the average human cell has around 30,000.
According to John Glass, senior author and leader of the synthetic biology group at JCVI, identifying those seven additional genes took seven years of painstaking effort.
Co-lead author and JCVI scientist Lijie Sun constructed dozens of variant strains by systematically adding and removing genes, and then she and the other researchers would observe how those genetic changes affected cell growth and division.
Eventually, the years of work paid off and the team discovered a combination of genes that led to JCVI-Syn3A cells dividing into cells of more uniform shape and size.
Specialist equipment
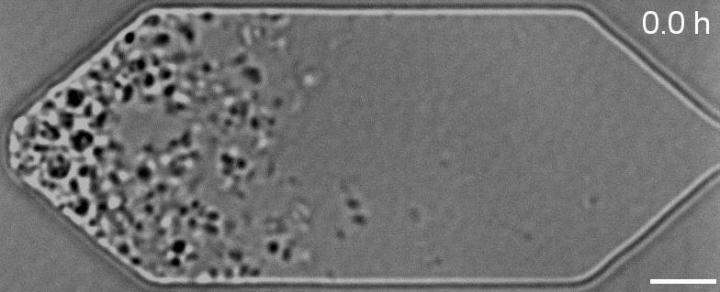
NIST’s role was to measure the resulting changes under a microscope, which was a challenge because the cells had to be alive for observation.
According to the researchers, using powerful microscopes to observe dead cells is relatively easy, but imaging live cells is much harder.
Holding these cells in place under a microscope was particularly difficult because they are so small and delicate; A hundred or more would fit inside a single E. coli bacterium and tiny forces can tear them apart.
To solve this problem, Strychalski and MIT co-authors James Pelletier, Andreas Mershin and Neil Gershenfeld designed a microfluidic chemostat – a sort of mini-aquarium – where the cells could be kept fed and happy under a light microscope.
The result was a stop-motion video that showed the synthetic cells growing and dividing, allowing the researchers to observe how their genetic manipulations affected the cell growth and division.
Does this answer all of the research team’s questions on synthetic cell division?
Although the breakthrough made by the research team helped illuminate the combination of genes required for normal cell division in a synthetic cell – there are still questions regarding the other genes added to make JCVI-syn3A.
Of the seven genes added to this organism for normal cell division, the scientists know what only two of them do; the roles that the other five play are not yet known, and will be their next focus,
“Our goal is to know the function of every gene so we can develop a complete model of how a cell works,” Pelletier said.
“Life is still a black box,” Strychalski added, but with this simplified synthetic cell, scientists are getting a good look at what’s going on inside.




